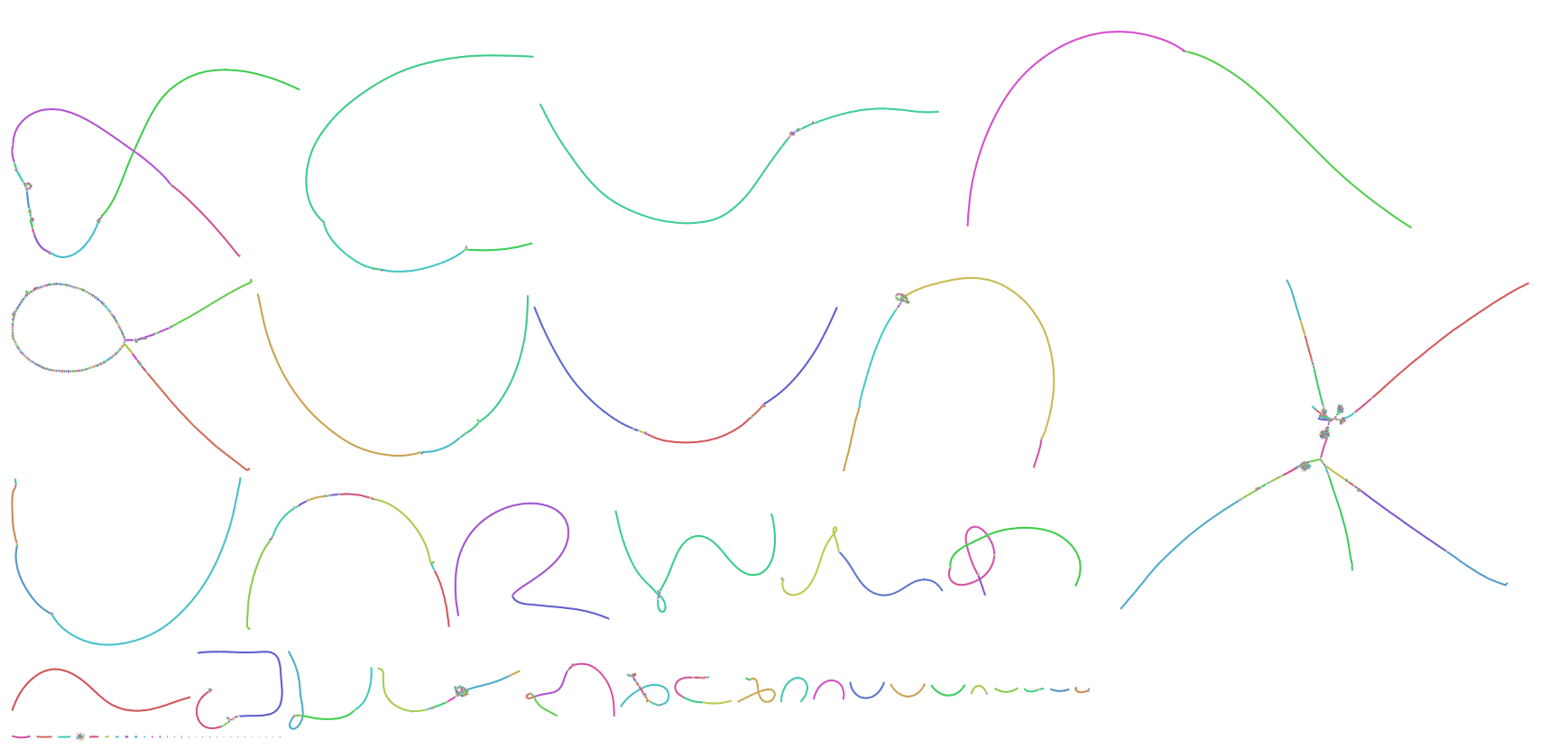Adam Phillippy | Genome Informatics Section | September 22, 2020
The Telomere-to-Telomere (T2T) consortium is proud to announce our v1.0 assembly of a complete human genome. This post briefly summarizes our work over the past year, including a month-long virtual workshop in June, as we strove to complete as many human chromosomes as possible. Our progress over the summer exceeded our wildest expectations and resulted in the completion of all human chromosomes, with the only exception being the 5 rDNA arrays. Our v1.0 assembly includes more than 100 Mbp of novel sequence compared to GRCh38, achieves near-perfect sequence accuracy, and unlocks the most complex regions of the genome to functional study. We plan to release a series of preprints in the coming months that fully describe our methods and analyses, but due to its tremendous value, we are releasing the assembly immediately.
Roughly twenty years ago, the International Human Genome Sequencing Consortium published an “Initial Sequencing and Analysis of the Human Genome” simultaneously with “The Sequence of the Human Genome” from Celera Genomics. Although the public consortium chose a more humble title that suggested some work was left to be done, amidst all the pomp it was easy to miss the fact that the human genome had not actually been finished. The key caveat appears early on in both papers, e.g. “A 2.91-billion base pair (bp) consensus sequence of the euchromatic portion of the human genome was generated”. Ignoring heterochromatin, due to difficulty in mapping, cloning, or assembling these sequences, excluded upwards of 10% of the genome from these initial drafts, and that missing fraction has been underappreciated ever since. Today, the latest human genome reference (GRCh38) still contains 161 Mbp of “unknown” sequence constituting 5% of the genome.
Now, twenty years later, we are finally able to fill in the blanks thanks to a confluence of new sequencing technologies from PacBio and Oxford Nanopore. Within the past year, the T2T consortium assembled the first complete human chromosomes, Chromosome X and Chromosome 8, using Nanopore ultra-long (UL) sequencing as a backbone and polishing that sequence with PacBio and Illumina. However, the recent release of PacBio’s HiFi technology led us to revise our recipe. In the assembly presented here, we first constructed a highly-accurate assembly graph using PacBio HiFi reads and then resolved any structural ambiguities with the help of Nanopore UL reads. The following image shows a Bandage visualization of our HiFi string graph for the CHM13 genome, with most chromosomes resolved as individual components.