
20 years after its debut, UCSC Genome Browser critical to pandemic-era research
Researchers worldwide using UC Santa Cruz’s “one-stop shop” for visualizing SARS-CoV-2 genome mutations
SANTA CRUZ, CA – September 9, 2020 – With campus COVID testing capacity at hundreds of tests a day and the start of the online fall quarter just around the corner, UC Santa Cruz Genomics Institute’s paper discussing its much-hailed coronavirus assembly browser has been published in Nature Genetics.
The paper gives an overview of the 30 kbp SARS-CoV-2 genome and the proteins it encodes. It details the supporting toolkit designed to promote accurate scientific inference and discourse.
In May, the New York Times’s Carl Zimmer tweeted that UCSC’s SARS-CoV-2 Genome Browser is a “one-stop shopping experience for #covid19 molecular biology.” The SARS-CoV-2 Browser is now getting thousands of hits per day.
NYT reporter Carl Zimmer tweeted that the Browser “a one-stop shopping experience for Covid-19 molecular biology.”
In late February, before it was declared a global threat, the then-novel coronavirus SARS-CoV-2 became the target of UC Santa Cruz’s genomics research team. On February 27, some of the same UC researchers who launched the Genome Browser 20 years ago unveiled the UCSC SARS-CoV-2 Genome Browser, marking the start of UCSC’s campaign against the virus.
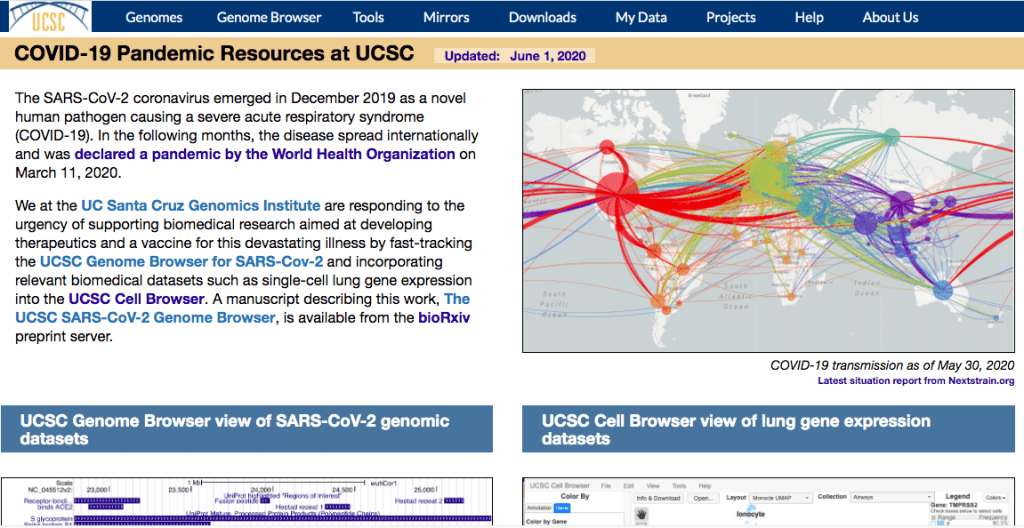
Globally, scientific investigation of the COVID-19 pandemic has generated what amounts to a mountain of molecular data pertaining to the SARS-CoV-2 RNA genome and its proteins.
The UCSC SARS-CoV-2 Browser serves a critical need “for rapid and continuously updated access to the latest molecular data in a format in which all data can be quickly cross-referenced and compared,” corresponding author Max Haeussler explained.
“This is a community resource: Anybody can add manual annotations which are quality checked and shared publicly on the browser the next day,” he added.
Hiram Clawson, the paper co-author who helped introduce the SARS-CoV-2 Browser this past winter says, “The UCSC Genome Browser is an established and highly accessed web-based viewer and standardized repository of genomic data with extensive functionality and a 20-year track record of serving the genomics community.”

Hiram Clawson of the UCSC Genome Browser team (top) unveils the UCSC SARS-CoV-2 Genome Browser on Feb. 27, marking the start of our fight against the virus.
New features including a “Crowd-Sourced Data” track enabling users to simply enter the coordinates and name of a feature, thus making the SARS-CoV-2 browser a simple but powerful tool to track developments in SARS-CoV-2 science, to detail the changes in the viral genome over the course of the pandemic, and to develop testable hypotheses and novel strategies to combat it.
Responding to limited coronavirus testing availability, affiliated researchers at the UC Santa Cruz Genomics Institute have also established the UCSC Molecular Diagnostic Lab, which began testing for the UCSC Student Health Center and local providers like Santa Cruz Community Health on May 1.
Both the Genome Browser and the UCSC Cell Browser are funded by the NIH National Human Genome Research Institute. UCSC COVID-19 projects are also funded by generous supporters, including Pat & Rowland Rebele; Eric and Wendy Schmidt, by recommendation of the Schmidt Futures program; the UC Center for Information Technology Research in the Interest of Society (CITRIS); and the University of California Office of the President (UCOP), as well as anonymous donors.

For more about The UCSC SARS-CoV-2 Genome Browser, see Dr. Haussler’s presentation given at HHMI Laboratories” virtual scientific meeting, “Confronting SARS-CoV-2,” held September 15-16, 2020.
Click here to access presentation.